Clinical implementation of rapid CYP2C19 genotyping to guide antiplatelet therapy after percutaneous coronary intervention
Abstract
Background
The CYP2C19 nonfunctional genotype reduces clopidogrel effectiveness after percutaneous coronary intervention (PCI). Following clinical implementation of CYP2C19 genotyping at University Florida (UF) Health Shands Hospital in 2012, where genotype results are available approximately 3 days after PCI, testing was expanded to UF Health Jacksonville in 2016 utilizing a rapid genotyping approach. We describe metrics with this latter implementation.
Methods
Patients at UF Health Jacksonville undergoing left heart catheterization with intent to undergo PCI were targeted for genotyping using the Spartan RX™ system. Testing metrics and provider acceptance of testing and response to genotype results were examined, as was antiplatelet therapy over the 6 months following genotyping.
Results
In the first year, 931 patients, including 392/505 (78%) total patients undergoing PCI, were genotyped. The median genotype test turnaround time was 96 min. Genotype results were available for 388 (99%) PCI patients prior to discharge. Of 336 genotyped PCI patients alive at discharge and not enrolled in an antiplatelet therapy trial, 1/6 (17%) poor metabolizers (PMs, with two nonfunctional alleles), 38/93 (41%) intermediate metabolizers (IMs, with one nonfunctional allele), and 119/237 (50%) patients without a nonfunctional allele were prescribed clopidogrel (p = 0.110). Clopidogrel use was higher among non-ACS versus ACS patients (78.6% vs. 42.2%, p < 0.001). Six months later, among patients with follow-up data, clopidogrel was prescribed in 0/4 (0%) PMs, 33/65 (51%) IMs, and 115/182 (63%) patients without a nonfunctional allele (p = 0.008 across groups; p = 0.020 for PMs versus those without a nonfunctional allele).
Conclusion
These data demonstrate that rapid genotyping is clinically feasible at a high volume cardiac catheterization facility and allows informed chronic antiplatelet prescribing, with lower clopidogrel use in PMs at 6 months.
Trial registration ClinicalTrials.gov Identifier: NCT02724319; registered March 31, 2016; https://www.clinicaltrials.gov/ct2/show/NCT02724319?term=angiolillo&rank=7
Background
Dual antiplatelet therapy with aspirin and a P2Y12 receptor inhibitor is the standard of care for patients with coronary artery disease (CAD) undergoing percutaneous coronary intervention (PCI) [1, 2]. Currently, three oral P2Y12 receptor antagonists (clopidogrel, prasugrel, and ticagrelor) are clinically available. Clopidogrel remains broadly utilized, which may be attributed to its lower cost, given that it is available in a generic formulation, and its expanded indications compared with prasugrel and ticagrelor [3, 4]. Prasugrel and ticagrelor in fact have an indication only for the treatment of patients with acute coronary syndrome (ACS), while clopidogrel is the only oral P2Y12 receptor antagonist also with an indication for the treatment of stable CAD [5]. Compared to clopidogrel, prasugrel and ticagrelor lead to a greater reduction in atherothrombotic events, albeit at the expense of increased bleeding not related to coronary artery bypass grafting, but clopidogrel is still frequently used [3, 4, 6].
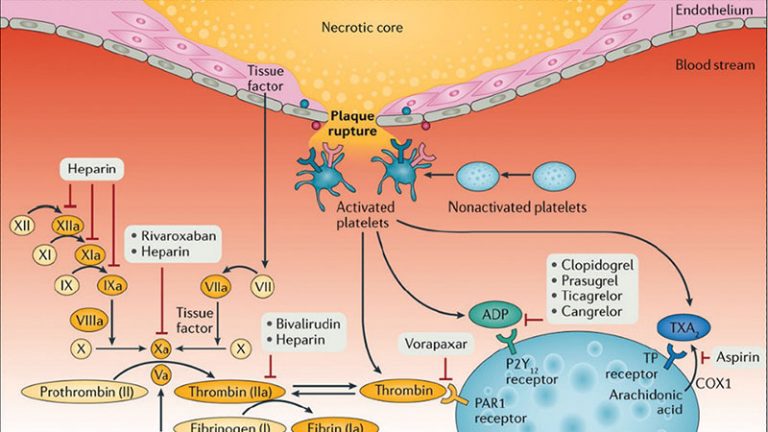
Clopidogrel is a thienopyridine that requires bioactivation to an active metabolite that irreversibly binds the platelet P2Y12 receptor and inhibits platelet activation and subsequent aggregation [3, 5]. There is significant inter-patient variability in clopidogrel-induced antiplatelet effects, which has important prognostic implications [7, 8]. In particular, in patients undergoing PCI, studies have consistently shown that those with reduced clopidogrel-induced antiplatelet effects, who thus persist with high on-clopidogrel platelet reactivity, are at high risk for ischemic recurrences, including stent thrombosis [8,9,10,11]. Emerging studies have also shown that the presence of enhanced clopidogrel-induced antiplatelet effects, leading to low platelet reactivity, may increase the risk of bleeding complications [9]. Multiple factors, including clinical, cellular, and genetic factors, may contribute to interindividual response variability to clopidogrel [7]. Among the genetic factors, polymorphisms in the CYP2C19 gene have consistently shown to have a role [12,13,14].
The CYP2C19 enzyme is involved in both metabolic steps mediating the biotransformation of clopidogrel to its pharmacologically active form. The CYP2C19*2 and *3 alleles are referred to as nonfunctional alleles and confer absent enzyme activity. These alleles are associated with lower plasma concentrations of the active metabolite and reduced platelet inhibition with clopidogrel [15]. Studies conducted in clopidogrel-treated patients undergoing PCI have shown an increased risk for ischemic events, in particular stent thrombosis, in the presence of a nonfunctional allele [12, 16]. Prasugrel and ticagrelor are newer P2Y12 receptor inhibitors that are not affected by the CYP2C19 genotype [17, 18], and consortium guidelines recommend consideration of these agents in patients with a nonfunctional genotype [14].
In June 2012, CYP2C19 testing on a GenMark Dx® platform (GenMark Diagnostics, Inc., Carlsbad, CA) was launched at the University of Florida (UF) Health Shands Hospital in Gainesville to assist with selection of antiplatelet therapy after PCI, and metrics with this implementation have been described [19]. This was a pharmacist-led effort. In April 2016, in a physician-led effort, CYP2C19 testing was launched at UF Health Jacksonville, whereby patients undergoing left heart catheterization with intent to undergo PCI were genotyped on a Spartan RX™ platform (Spartan Bioscience Inc, Ottawa, ON). This provided experience with a rapid genotyping approach to clinical CYP2C19 testing. Herein we describe implementation metrics associated with CYP2C19 genotype-guided antiplatelet therapy at UF Health Jacksonville. We specifically report on physician uptake of CYP2C19 testing, genotype results and turnaround time, and both acute and chronic antiplatelet therapy prescribed after genotype results were available. We also describe lessons learned based on our experiences with two approaches to CYP2C19 genotyping implementation.
Methods
Procedures for implementation
Patients undergoing emergent or planned left heart catheterization with intent to undergo PCI were targeted for testing. Patients with a history of human immunodeficiency virus or hepatitis C virus were excluded. Written informed consent for clinical CYP2C19 testing and collection of clinical data and a blood sample for future research were obtained from each patient. Genetic samples were collected via a buccal swab prior to cardiac catheterization when possible. For patients undergoing emergency procedures (e.g., ST-segment elevation myocardial infarction) or otherwise unable to provide consent prior to catheterization (e.g., patients with cardiogenic shock or intubated), informed consent and genetic sampling were obtained following PCI after patients were stabilized. Patients not receiving PCI were not tested after diagnostic left heart catheterization.
Genetic samples were transported to the college of American pathologist/CLIA licensed pathology laboratory, located five floors below the cardiac catheterization laboratory. Samples were processed using the Spartan RX™ system, which tests for the CYP2C19 nonfunctional *2 (c.681G>A; rs4244285) and *3 (c.636G>A; rs4986893) alleles and the increased function *17 (c.-806C>T; rs12248560) allele [20]. Those lacking a *2, *3, or *17 allele were assigned the *1 allele designation. Phenotypes were assigned according to standardized nomenclature, with PMs having two nonfunctional alleles (e.g. *2/*2), IMs having a single nonfunctional allele (e.g. *1/*2, *2/*17), normal metabolizers (NMs) having the *1/*1 genotype, and rapid and ultra-rapid metabolizers (RMs and UMs) having the *1/*17 and *17/*17 genotypes, respectively [21]. One Spartan RX™ system was initially available, but a second system was added in May 2017 to accommodate testing multiple patients at once. In the event of inconclusive genotype results, collection of an additional buccal cell sample for genotyping was attempted. Testing was only available Monday through Friday. For patients presenting on the weekend for emergency PCI, samples were collected on Monday for genotyping if the patient remained hospitalized.
The overall goal was to have genetic information available by the end of the PCI procedure or before discharge for patients unable to provide informed consent prior to coronary intervention. Genotype and phenotype results were placed in the Epic electronic health record (EHR) under the laboratory reports tab, with phenotype assignment as described above. Results were communicated to physicians via an inbox message to use at their discretion taking into consideration other factors (e.g., clinical presentation, cardiovascular risk factors, concomitant medications, insurance, PCI complexity) in making antiplatelet therapy decisions. No clinical pharmacist support was provided. Genotyping kits and systems were donated by Spartan Biosciences, Inc. Technical and pathologist time was paid through grant support.
Data collection and analysis
Data for the first 12 months of implementation were collected on the genotype test ordering rate, genotype turnaround time (defined as the time between the sample arrival in the pathology laboratory to the time the report was signed by the pathologist) and genotype results. For patients receiving PCI, data were also collected on antiplatelet therapy at discharge and 6 months after coronary intervention. Metrics data were tracked through review of patient encounters in the EHR and telephone calls. Use of clopidogrel was compared between PMs, IMs, and patients without a nonfunctional allele (e.g., combination of NMs, RMs, and UMs) using Chi square analysis or the Fisher’s exact test. Changes in antiplatelet therapy from baseline to 6 months were assessed within each phenotype group using the McNemar test. All analyses were conducted using SAS version 9.4 (SAS, Cary, NC). Study procedures and data collection were approved by the Institutional Review Board at UF Health Jacksonville, and all procedures were in accordance with the ethical standards of the Declaration of Helsinki.
Results
From April 28, 2016 to April 27, 2017, 931 patients undergoing left heart catheterization for suspected coronary disease were genotyped, representing 68% (931/1366) of all patients undergoing left heart catheterization during that period (Table 1). The median genotype test turnaround time was 96 (interquartile range of 78–144) min. Genotyping for 129 patients (14%) was unsuccessful with the initial sample (56 inconclusive results, 73 device errors). One additional sample was collected in 113 patients, two additional samples were collected in 10 patients, and 6 patients refused sample recollection. Nine of the 123 patients with additional sample collection had multiple inconclusive results.
Table 1 CYP2C19 test ordering and adoption rates in the first 12 months at UF Health JacksonvilleFull size table
A total of 392 of the 931 genotyped patients (42%) underwent PCI, representing 78% (392/505) of all patients undergoing PCI during the 12 month period. Of these, 4 (1%) had inconclusive genotype results. Of the 113 patients receiving PCI and not genotyped, 12 refused to participate in the study and the remaining 101 were excluded, unable to provide informed consent, or not approached about testing (e.g. underwent PCI over the weekend and were discharged before they could be consented). The characteristics of the genotyped patients receiving PCI are shown in Table 2. The majority were male and presented with an ACS at the time of PCI. Twenty-nine percent had a *2 allele; 27% were intermediate metabolizers (IMs) and 2% were poor metabolizers (PMs). The *3 allele was not detected in any patient.
Table 2 Characteristics of patients who underwent PCI and CYP2C19 genotyping over the first 12 monthsFull size table
Fifty-one patients (14 with a *2 allele and 37 without a *2 allele) were enrolled in the Ticagrelor With Aspirin or Alone in High-Risk Patients After Coronary Intervention (TWILIGHT) trial (ClinicalTrials.gov Identifier: NCT02270242), which dictated treatment with ticagrelor for 3 months following PCI [22]. An additional patient died prior to hospital discharge. After excluding these 52 patients, plus 4 patients with inconclusive genotype results, 1 of 6 (17%) PMs, 38 of 93 (41%) IMs, and 119 of 237 patients without a *2 allele (50%) were placed on clopidogrel at the time of discharge (p = 0.110 for comparison across groups; Fig. 1). The remaining 178/336 patients were treated with prasugrel (81/178, 45.5%) or ticagrelor (97/178, 54.5%).
Fig. 1
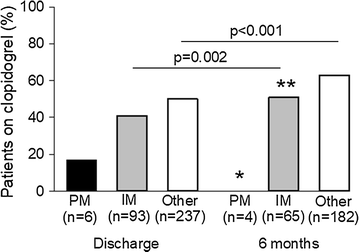
Percent of patients on clopidogrel at discharge and 6 months by CYP2C19 phenotype. PM poor metabolizer, IM intermediate metabolizer. Other includes normal metabolizers, rapid metabolizers, and ultra-rapid metabolizers. *p = 0.020 for PM versus other at 6 months. **p = 0.104 for IM versus other at 6 monthsFull size image
At the time of data collection for the present analysis, 6 month post-PCI follow-up data were available for 258 patients. Of 251 still taking a P2Y12 inhibitor, use of clopidogrel increased significantly in IMs (p = 0.002) and patients without a *2 allele (p < 0.001) from discharge to 6 months, but remained low in PMs. At 6 months, 0/4 (0%) PMs, 33/65 (51%) IMs, and 115/182 (63%) patients without a *2 allele were prescribed clopidogrel (p = 0.008 for comparison across groups). Clopidogrel use was significantly higher in those without a *2 allele compared to PMs (p = 0.020), but not compared to IMs (p = 0.104).
When stratifying antiplatelet therapy by PCI indication, there was significantly higher use of clopidogrel in non-ACS patients (n = 28) versus ACS patients (n = 223) at discharge (78.6% vs. 42.2%, p < 0.001) and 6 months (82.1% vs. 56.1%, p = 0.008). Similar to data in the whole population, among ACS patients, clopidogrel use increased significantly from baseline to 6 months in patients without a CYP2C19*2 allele (45.6% at discharge to 58.9% at 6 months, p < 0.001), as well as in IMs (35.5% at discharge to 51.6% at 6 months, p = 0.004), whereas all PMs received alternative therapy at discharge and 6 months. There were too few non-ACS patients for analysis of changes in antiplatelet therapy over time by genotype.
Lessons learned
The implementation at UF Health Shands Hospital in Gainesville has been previously described [19]. Table 3 compares implementation approaches between UF Health Jacksonville and UF Health Shands Hospital in Gainesville and lessons learned based on experience with the two approaches. A major challenge with the genotyping approach in Gainesville was that the delay in obtaining genotype results significantly disrupted provider workflow given the need to follow up on results returned 3–4 days after the PCI procedure, which was often after the patient had been discharged. An effective communication strategy utilizing both clinical pharmacist support and electronic clinical decision support was necessary in this setting to alert providers about patients with a nonfunctional allele so that appropriate antiplatelet therapy could be instituted. This approach was successful in ensuring high uptake of prasugrel or ticagrelor in both PMs and IMs, whereas most patients without a nonfunctional allele were treated with clopidogrel. In contrast, neither pharmacist support nor clinical decision support were used to assist with antiplatelet therapy in Jacksonville where results are usually available 1–2 h after sample collection, and well before patient discharge. Also in contrast to UF Health, Gainesville, prasugrel or ticagrelor was commonly prescribed in the acute post-PCI phase at UF Health, Jacksonville regardless of genotype. Genotype was available to inform switches after the acute phase from more potent and expensive P2Y12 inhibitors to clopidogrel in patients without a nonfunctional allele, as evidenced by significantly increased use of clopidogrel in those without a *2 over the initial 6 month period and continued use of prasugrel or ticagrelor in PMs. Interestingly, use of clopidogrel in IMs increased over time at UF Health, Jacksonville, which is against guideline recommendations. Thus, an important lessons learned is that some sort of clinical decision support is needed to assist with genotype-guided chronic antiplatelet therapy decisions. Another lesson learned was that genotyping was useful in informing antiplatelet therapy in the setting where clopidogrel was the preferred antiplatelet agent (at UF Health, Gainesville) and in the setting where more potent P2Y12 inhibitors were preferred early after PCI (at UF Health, Jacksonville), with a switch to clopidogrel after the acute period, assuming that clinical decision support can be put in place to ensure appropriate genotype-guided clopidogrel use.Table 3 Implementation approaches at two UF Health locations and lessons learnedFull size table
Discussion
We previously showed that CYP2C19 testing in conjunction with clinical decision support and clinical pharmacist participation can be successfully integrated into clinical care to guide antiplatelet therapy after PCI, with high prescriber adoption rates of the testing and recommendations for alternative therapy for patients with a nonfunctional allele during the initial year of the program [19]. Herein, we extend those findings based on a physician-led implementation at another UF Health institution. While there were some research aspects to the implementation, including obtaining consent for sample storage for future research, most aspects of the implementation were done as part of clinical care, with genotyping performed in a CAP/CLIA-licensed laboratory, results placed in the EHR, and prescribing decisions left to the provider. Other institutions across the United States have also clinically implemented CYP2C19 testing to guide post-PCI antiplatelet therapy, with a description of strategies undertaken recently published [23]. Consistent with a recent publication from the University of Alabama, Birmingham [24], we show that, in the largest single institution study reported to date, use of a rapid CYP2C19 genotyping platform outside of a clinical trial is clinically feasible. We further show that the availability of a rapid and user-friendly genotyping platform led to high adoption of CYP2C19 testing for patients presenting to the cardiac catheterization laboratory with intent for PCI.
Clopidogrel reduces the risk for adverse cardiovascular events after an ACS, and additional data show reductions in the risk for cardiovascular events, including stent thrombosis, with clopidogrel use after PCI [1, 5, 6]. However, the CYP2C19 nonfunctional genotype compromises the efficacy of clopidogrel, leading to reduced formation of the active thiol metabolite and reduced inhibition of platelet aggregation [15]. While the impact of CYP2C19 genotype on clopidogrel efficacy in lower risk populations who do not undergo PCI is questionable [25, 26], the data consistently show reduced clopidogrel effectiveness after PCI in carriers of a nonfunctional allele, with the lowest effectiveness observed in PMs [12, 16]. Based on these data, guidelines by the Clinical Pharmacogenetics Implementation Consortium (CPIC) recommend integrating available genotype results into prescribing decisions for antiplatelet therapy after PCI [14]. They specifically recommend prasugrel or ticagrelor for patients with a LOF allele in the absence of contraindications.
While CPIC guidelines do not provide recommendations on whether or not to order genotyping, leaving that to the discretion of the physician, this is addressed in practice guidelines for the management of patients undergoing PCI [1, 2]. These guidelines recommend against routine use of genetic testing for all patients undergoing PCI, citing an absence of data from large randomized controlled trials, but state that testing may be considered for patients at high risk for poor outcomes. Two multi-center trials assessing the efficacy of genotype-guided antiplatelet prescribing after PCI are ongoing but are not expected to be completed for at least a year (ClinicalTrials.gov Identifiers: NCT01742117 and NCT01761786). There are, however, recent data on outcomes from a pragmatic clinical trial showing a reduced risk of the composite of ischemic and bleeding events with genotype-guided antiplatelet therapy versus usual care in patients with ACS, most of whom underwent PCI [27]. There are additional data from pragmatic studies of CYP2C19 genotyping in clinical care. Specifically, we showed in a study of 412 patients who underwent CYP2C19 genotyping and PCI at UF Health in Gainesville that there was a higher incidence of major adverse cardiovascular events (MACE), consisting of cardiovascular death, myocardial infarction, stroke, or stent thrombosis, in patients with a nonfunctional allele treated with clopidogrel versus prasugrel or ticagrelor [28]. In a subsequent study of 1815 patients genotyped across seven medical centers, we observed an increased risk for MACE, defined as death from any cause, non-fatal myocardial infarction, or non-fatal stroke, in nonfunctional allele carriers treated with clopidogrel versus alternative therapy (adjusted hazard ratio of 2.26, 95% confidence interval 1.18–4.32) [29]. There remained significantly worse outcomes with clopidogrel compared to alternative therapy when limiting the analysis to IMs. This is noteworthy because the FDA-approved clopidogrel labeling includes a boxed warning about reduced drug effectiveness in PMs, with two nonfunctional alleles, but does not address IMs, with a single nonfunctional allele [30]. A cost-effectiveness analysis based on event rates in this study is underway. Previous cost-effectiveness analyses using event rate probabilities derived from clinical trials or cohort studies showed that a genotype-guided approach to antiplatelet therapy after ACS and PCI is cost effective compared to universal clopidogrel, ticagrelor, or prasugrel [31, 32].
There were a number of valuable lessons learned based on our experience implementing CYP2C19 testing at two different hospitals within UF Health. First, a number of conditions must be met for rapid genotyping to be feasible in the clinical setting. A genetic test is considered a high complexity test according to the Clinical Laboratory Improvement Amendments (CLIA) and must be performed according to a college of American pathologist/CLIA accredited process in the US [33]. Thus, while rapid genotyping platforms may be designed as point-of-care tests outside the US, they cannot be used in this manner in the US in the absence of a licensed molecular medical technologist available to perform the test. This generally requires the platform to be placed in the CAP/CLIA certified pathology laboratory, as done at UF Health Jacksonville. Relative to the Spartan RX™ system, the only FDA-cleared platform for rapid CYP2C19 genotyping, the genetic sample must be placed in the instrument within an hour of collection, necessitating dedicated staff to efficiently deliver the sample to the pathology laboratory. While this process works well at UF Health Jacksonville given the close proximity of the cardiac catheterization and clinical pathology laboratories, it is not feasible at UF Health Shands Gainesville where the pathology laboratory is located farther away. Another limitation of the Spartan RX™ system is that only a single sample can be genotyped at a time. Thus, to increase genotyping capacity at the Jacksonville site, a second system was added. However, this still limited the number of patients we could genotype and contributed to our inability to offer genotyping to all patients undergoing left heart catheterization. Finally, 13.9% of samples initially failed to genotype with the Spartan RX™ system, requiring repeat sample collection and increasing the cost of testing.
The approach taken at UF Health Jacksonville was to genotype all patients undergoing left heart catheterization with intent for PCI. While 58% of patients did not immediately proceed to PCI, in the event that patients return for PCI in the future, genotype will be readily available in the EHR to help inform therapy. The availability of a rapid genotyping platform, however, negates the need to genotype all patients at the time of cardiac catheterization and allows for efficient genotyping at the time of PCI.
Another important lesson is that when genotype results are not available for several days after PCI, as was the case at UF Health Gainesville, it creates significant disruptions in workflow caused by the need to adjust therapy after the patient has left the cardiac catheterization laboratory and potentially after the patient has been discharged from the hospital. In fact, this was the primary barrier cited by two large private practices in Florida with high volume cardiac catheterization laboratories, who we previously approached about implementing CYP2C19 testing. This suggests that to implement testing more broadly, genotyping should cause minimal to no negative impact on work flow. Solutions to this problem are either preemptive pharmacogenetic testing, so that results are available ahead of PCI, or rapid genotyping at the time of PCI as done at UF Health Jacksonville. In the landscape of a lack of reimbursement for preemptive pharmacogenetic testing, the use of rapid genotyping platforms may be the most feasible solution at this time.
A third lesson learned was that despite the shift to newer agents early after ACS and PCI, genotype still has an important role in informing on the choice of long-term antiplatelet therapy and specifically with informing de-escalation from more potent antiplatelet therapy early after PCI to clopidogrel for maintenance therapy. This has important prognostic implications given that most ischemic events occur early (i.e., first month), while bleeding events continue to accrue over time particularly with the long-term use of prasugrel and ticagrelor [34, 35]. These observations have led to experts to advocate that the use of prasugrel or ticagrelor be limited to the first few weeks or month after PCI to reduce the risk of ischemic events, and then therapy de-escalated to clopidogrel to minimize the risk of bleeding complications [3, 36]. However, a non-guided de-escalation approach (e.g. without genotyping or platelet function testing) to a less potent agent has been associated with conflicting outcomes findings [37, 38], with very early de-escalation after an ACS event associated with an increased risk of ischemic events. On the contrary, the recently reported testing responsiveness to platelet inhibition on chronic antiplatelet treatment for acute coronary syndromes (TROPICAL-ACS) trial, using a platelet function guided approach, provides a justification for a de-escalation strategy [39]. The trial specifically demonstrated that de-escalation from prasugrel to clopidogrel, guided by platelet function testing, was non-inferior to continued prasugrel use in preventing adverse cardiovascular events after ACS and PCI. A limitation with platelet function testing is that it must be performed while the patient is on treatment. That is, patients in the TROPICAL-ACS trial were switched to clopidogrel for 1 week prior to testing, and only those with sufficient evidence of platelet inhibition were continued on clopidogrel, whereas those with high on-treatment platelet reactivity were switched back to prasugrel. A strategy of de-escalation and escalation back to prasugrel or ticagrelor with repeated measures of platelet function may not always be practical and can be inconvenient for the patient [40]. Genotyping may represent another means of guiding de-escalation, and unlike platelet function testing, it can be done apriori, without the need to switch the patient to clopidogrel prior to testing.
In the current study, we observed continued high use of prasugrel or ticagrelor in CYP2C19 PMs, who have significantly limited capacity to activate clopidogrel. However, in line with a genotype-guided de-escalation approach, use of alternative antiplatelet therapy declined significantly over the 6 month period after PCI among those without a nonfunctional allele. Interestingly, there was a similar decline in IMs. The preference for continuing alternative therapy in PMs, but not necessarily in IMs, could be based on the boxed warning on the FDA-approved clopidogrel labeling, which is specific to PMs [30]. Data from the pragmatic study described above showing improved outcomes in IMs treatment with alternative therapy versus clopidogrel support continuation of alternative therapy in IMs [29]. However, these data were not available during the first year of implementation at UF Health Jacksonville. Of note, a limitation of our findings is that we did not specifically assess whether factors, other than genotype, contributed to decisions to switch antiplatelet therapy, such as patient ability to afford alternative agents and presence of conditions that increase bleeding risk (e.g., use of concomitant oral anticoagulants, frailty, and history of intracranial or other serious bleeding), nor did we collect data on loading doses or on patients not undergoing genotyping for comparison.
An additional lesson learned was the importance of clinical pharmacy support and electronic clinical decision support when there are delays in obtaining genotype results to assist with communicating results to providers after the patient has left the cardiac catheterization laboratory and in some cases, after the patients has been discharged from the hospital. In settings where rapid genotyping is available and initial use of alternative therapy post PCI is high, one could argue that pharmacist support may not be necessary to assist with early antiplatelet treatment decisions. Nonetheless, approximately 40% of IMs in the current study were placed on clopidogrel acutely, which is inconsistent with CPIC guidelines, suggesting that clinical pharmacist support and/or electronic clinical decision support may be warranted regardless of genotype turnaround time. Pharmacist support and electronic decision support may also be important in the chronic management of patients to assist genotype-guided decisions to de-escalate antiplatelet therapy to clopidogrel. Indeed, at UF Health, Gainesville, where there is significant pharmacist support plus electronic decision support, the majority of IMs were switched from clopidogrel to prasugrel or ticagrelor, consistent with CPIC recommendations. However, the opposite occurred in the absence of such support at UF Health, Jacksonville. Pharmacists can also play important roles in educating providers and patients on genetic test results and implications for treatment response regardless of genetic testing procedures.
Conclusion
Our data show that providing genotype-guided antiplatelet therapy after PCI with the use of a rapid genotyping platform is feasible with high provider acceptance of genetic testing. This is among the first reports of implementation metrics with clinical use of a rapid CYP2C19 genotyping platform and the largest study of rapid CYP2C19 genotyping in clinical care to date. With increasing use of newer antiplatelet agents, especially in the setting of ACS, our data suggest that genotyping may have an important role in informing maintenance antiplatelet therapy following the early post-PCI period when risk for adverse events is highest.
References
- 1.Levine GN, Bates ER, Blankenship JC, Bailey SR, Bittl JA, Cercek B, Chambers CE, Ellis SG, Guyton RA, Hollenberg SM, et al. 2015 ACC/AHA/SCAI focused update on primary percutaneous coronary intervention for patients with ST-elevation myocardial infarction: an Update of the 2011 ACCF/AHA/SCAI guideline for percutaneous coronary intervention and the 2013 ACCF/AHA guideline for the management of ST-elevation myocardial infarction: a report of the American college of cardiology/American heart association task force on clinical practice guidelines and the society for cardiovascular angiography and interventions. Circulation. 2016;133:1135–47.Article PubMed Google Scholar
- 2.Valgimigli M, Bueno H, Byrne RA, Collet JP, Costa F, Jeppsson A, Juni P, Kastrati A, Kolh P, Mauri L, et al. 2017 ESC focused update on dual antiplatelet therapy in coronary artery disease developed in collaboration with EACTS: the task force for dual antiplatelet therapy in coronary artery disease of the European society of cardiology (ESC) and of the European association for cardio-thoracic surgery (EACTS). Eur Heart J. 2017;53(1):34–78.Google Scholar
- 3.Rollini F, Franchi F, Angiolillo DJ. Switching P2Y12-receptor inhibitors in patients with coronary artery disease. Nat Rev Cardiol. 2016;13:11–27.CAS Article PubMed Google Scholar
- 4.Fan W, Plent S, Prats J, Deliargyris EN. Trends in P2Y12 inhibitor use in patients referred for invasive evaluation of coronary artery disease in contemporary US practice. Am J Cardiol. 2016;117:1439–43.CAS Article PubMed Google Scholar
- 5.Park Y, Franchi F, Rollini F, Angiolillo DJ. Dual antiplatelet therapy after coronary stenting. Expert Opin Pharmacother. 2016;17:1775–87.CAS Article PubMed Google Scholar
- 6.Franchi F, Angiolillo DJ. Novel antiplatelet agents in acute coronary syndrome. Nat Rev Cardiol. 2015;12:30–47.CAS Article PubMed Google Scholar
- 7.Angiolillo DJ, Fernandez-Ortiz A, Bernardo E, Alfonso F, Macaya C, Bass TA, Costa MA. Variability in individual responsiveness to clopidogrel: clinical implications, management, and future perspectives. J Am Coll Cardiol. 2007;49:1505–16.CAS Article PubMed Google Scholar
- 8.Tantry US, Bonello L, Aradi D, Price MJ, Jeong YH, Angiolillo DJ, Stone GW, Curzen N, Geisler T, Ten Berg J, et al. Consensus and update on the definition of on-treatment platelet reactivity to adenosine diphosphate associated with ischemia and bleeding. J Am Coll Cardiol. 2013;62:2261–73.CAS Article PubMed Google Scholar
- 9.Aradi D, Kirtane A, Bonello L, Gurbel PA, Tantry US, Huber K, Freynhofer MK, ten Berg J, Janssen P, Angiolillo DJ, et al. Bleeding and stent thrombosis on P2Y12-inhibitors: collaborative analysis on the role of platelet reactivity for risk stratification after percutaneous coronary intervention. Eur Heart J. 2015;36:1762–71.CAS Article PubMed Google Scholar
- 10.Stone GW, Witzenbichler B, Weisz G, Rinaldi MJ, Neumann FJ, Metzger DC, Henry TD, Cox DA, Duffy PL, Mazzaferri E, et al. Platelet reactivity and clinical outcomes after coronary artery implantation of drug-eluting stents (ADAPT-DES): a prospective multicentre registry study. Lancet. 2013;382:614–23.Article PubMed Google Scholar
- 11.Brar SS, ten Berg J, Marcucci R, Price MJ, Valgimigli M, Kim HS, Patti G, Breet NJ, DiSciascio G, Cuisset T, et al. Impact of platelet reactivity on clinical outcomes after percutaneous coronary intervention. A collaborative meta-analysis of individual participant data. J Am Coll Cardiol. 2011;58:1945–54.Article PubMed Google Scholar
- 12.Mega JL, Simon T, Collet JP, Anderson JL, Antman EM, Bliden K, Cannon CP, Danchin N, Giusti B, Gurbel P, et al. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA. 2010;304:1821–30.CAS Article PubMed PubMed Central Google Scholar
- 13.Moon JY, Franchi F, Rollini F, Rivas Rios JR, Kureti M, Cavallari LH, Angiolillo DJ. Role of genetic testing in patients undergoing percutaneous coronary intervention. Expert Rev Clin Pharmacol. 2018;11:151–64.CAS Article PubMed Google Scholar
- 14.Scott SA, Sangkuhl K, Stein CM, Hulot JS, Mega JL, Roden DM, Klein TE, Sabatine MS, Johnson JA, Shuldiner AR, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther. 2013;94:317–23.CAS Article PubMed PubMed Central Google Scholar
- 15.Mega JL, Close SL, Wiviott SD, Shen L, Hockett RD, Brandt JT, Walker JR, Antman EM, Macias W, Braunwald E, et al. Cytochrome p-450 polymorphisms and response to clopidogrel. N Engl J Med. 2009;360:354–62.CAS Article PubMed Google Scholar
- 16.Sorich MJ, Rowland A, McKinnon RA, Wiese MD. CYP2C19 genotype has a greater effect on adverse cardiovascular outcomes following percutaneous coronary intervention and in Asian populations treated with clopidogrel: a meta-analysis. Circ Cardiovasc Genet. 2014;7:895–902.CAS Article PubMed Google Scholar
- 17.Mega JL, Close SL, Wiviott SD, Shen L, Hockett RD, Brandt JT, Walker JR, Antman EM, Macias WL, Braunwald E, et al. Cytochrome P450 genetic polymorphisms and the response to prasugrel: relationship to pharmacokinetic, pharmacodynamic, and clinical outcomes. Circulation. 2009;119:2553–60.CAS Article PubMed Google Scholar
- 18.Wallentin L, James S, Storey RF, Armstrong M, Barratt BJ, Horrow J, Husted S, Katus H, Steg PG, Shah SH, et al. Effect of CYP2C19 and ABCB1 single nucleotide polymorphisms on outcomes of treatment with ticagrelor versus clopidogrel for acute coronary syndromes: a genetic substudy of the PLATO trial. Lancet. 2010;376:1320–8.CAS Article PubMed Google Scholar
- 19.Weitzel KW, Elsey AR, Langaee TY, Burkley B, Nessl DR, Obeng AO, Staley BJ, Dong HJ, Allan RW, Liu JF, et al. Clinical pharmacogenetics implementation: approaches, successes, and challenges. Am J Med Genet C Semin Med Genet. 2014;166C:56–67.Article PubMed Google Scholar
- 20.Roberts JD, Wells GA, Le May MR, Labinaz M, Glover C, Froeschl M, Dick A, Marquis JF, O’Brien E, Goncalves S, et al. Point-of-care genetic testing for personalisation of antiplatelet treatment (RAPID GENE): a prospective, randomised, proof-of-concept trial. Lancet. 2012;379:1705–11.CAS Article PubMed Google Scholar
- 21.Caudle KE, Dunnenberger HM, Freimuth RR, Peterson JF, Burlison JD, Whirl-Carrillo M, Scott SA, Rehm HL, Williams MS, Klein TE, et al. Standardizing terms for clinical pharmacogenetic test results: consensus terms from the clinical pharmacogenetics implementation consortium (CPIC). Genet Med. 2017;19:215–23.Article PubMed Google Scholar
- 22.Baber U, Dangas G, Cohen DJ, Gibson CM, Mehta SR, Angiolillo DJ, Pocock SJ, Krucoff MW, Kastrati A, Ohman EM, et al. Ticagrelor with aspirin or alone in high-risk patients after coronary intervention: rationale and design of the TWILIGHT study. Am Heart J. 2016;182:125–34.CAS Article PubMed Google Scholar
- 23.Empey PE, Stevenson JM, Tuteja S, Weitzel KW, Angiolillo DJ, Beitelshees AL, Coons JC, Duarte JD, Franchi F, Jeng LJB, et al. Multisite investigation of strategies for the implementation of CYP2C19 genotype-guided antiplatelet therapy. Clin Pharmacol Ther. 2017. https://doi.org/10.1002/cpt.1006.PubMed Google Scholar
- 24.Harada S, Zhou Y, Duncan S, Armstead AR, Coshatt GM, Dillon C, Brott BC, Willig J, Alsip JA, Hillegass WB, et al. Precision medicine at the university of Alabama at Birmingham: laying the foundational processes through implementation of genotype-guided antiplatelet therapy. Clin Pharmacol Ther. 2017;102:493–501.CAS Article PubMed Google Scholar
- 25.Doll JA, Neely ML, Roe MT, Armstrong PW, White HD, Prabhakaran D, Winters KJ, Duvvuru S, Sundseth SS, Jakubowski JA, et al. Impact of CYP2C19 metabolizer status on patients with ACS treated with prasugrel versus clopidogrel. J Am Coll Cardiol. 2016;67:936–47.CAS Article PubMed Google Scholar
- 26.Pare G, Mehta SR, Yusuf S, Anand SS, Connolly SJ, Hirsh J, Simonsen K, Bhatt DL, Fox KA, Eikelboom JW. Effects of CYP2C19 genotype on outcomes of clopidogrel treatment. N Engl J Med. 2010;363:1704–14.CAS Article PubMed Google Scholar
- 27.Notarangelo FM, Maglietta G, Bevilacqua P, Cereda M, Merlini PA, Villani GQ, Moruzzi P, Patrizi G, Tagliazucchi GM, Crocamo A, et al. Pharmacogenomic approach to selecting antiplatelet therapy in acute coronary syndromes: PHARMCLO trial. J Am Coll Cardiol. 2018. https://doi.org/10.1016/j.jacc.2018.02.029.PubMed Google Scholar
- 28.Cavallari LH, Magvanjav O, Anderson RD, Gong Y, Owusu-Obeng A, Kong B, Vo T, Ashton JN, Staley BJ, Elsey AR, et al. Clinical implementation of CYP2C19 genotype guided antiplatelet therapy reduces cardiovascular events after PCI. Circulation. 2015;132:A11802. http://circ.ahajournals.org/content/132/Suppl_3/A.abstract?sid=cd6bc466-3451-41a8-8339-503e71988d82.
- 29.Cavallari LH, Lee CR, Beitelshees AL, Cooper-DeHoff RM, Duarte JD, Voora D, Kimmel SE, McDonough CW, Gong Y, Dave CV, et al. Multi-site investigation of outcomes with implementation of CYP2C19 genotype-guided antiplatelet therapy after percutaneous coronary intervention. JACC Cardiovasc Interv. 2018;11:181–91.Article PubMed Google Scholar
- 30.Holmes DR Jr, Dehmer GJ, Kaul S, Leifer D, O’Gara PT, Stein CM. ACCF/AHA clopidogrel clinical alert: approaches to the FDA “boxed warning”: a report of the American college of cardiology foundation task force on clinical expert consensus documents and the American heart association endorsed by the society for cardiovascular angiography and interventions and the society of thoracic surgeons. J Am Coll Cardiol. 2010;56:321–41.CAS Article PubMed Google Scholar
- 31.Johnson SG, Gruntowicz D, Chua T, Morlock RJ. Financial analysis of CYP2C19 genotyping in patients receiving dual antiplatelet therapy following acute coronary syndrome and percutaneous coronary intervention. J Manag Care Spec Pharm. 2015;21:552–7.Article PubMed Google Scholar
- 32.Kazi DS, Garber AM, Shah RU, Dudley RA, Mell MW, Rhee C, Moshkevich S, Boothroyd DB, Owens DK, Hlatky MA. Cost-effectiveness of genotype-guided and dual antiplatelet therapies in acute coronary syndrome. Ann Intern Med. 2014;160:221–32.Article PubMed Google Scholar
- 33.Valdes R, Payne DA, Linder MW. Laboratory analysis and application of pharmacogenetics to clinical practice. Washington, D.C.: National Academy of Clinical Biochemistry; 2010.Google Scholar
- 34.Becker RC, Bassand JP, Budaj A, Wojdyla DM, James SK, Cornel JH, French J, Held C, Horrow J, Husted S, et al. Bleeding complications with the P2Y12 receptor antagonists clopidogrel and ticagrelor in the PLATelet inhibition and patient Outcomes (PLATO) trial. Eur Heart J. 2011;32:2933–44.CAS Article PubMed Google Scholar
- 35.Antman EM, Wiviott SD, Murphy SA, Voitk J, Hasin Y, Widimsky P, Chandna H, Macias W, McCabe CH, Braunwald E. Early and late benefits of prasugrel in patients with acute coronary syndromes undergoing percutaneous coronary intervention: a TRITON-TIMI 38 (TRial to Assess Improvement in Therapeutic Outcomes by Optimizing Platelet InhibitioN with Prasugrel-Thrombolysis In Myocardial Infarction) analysis. J Am Coll Cardiol. 2008;51:2028–33.CAS Article PubMed Google Scholar
- 36.Angiolillo DJ, Rollini F, Storey RF, Bhatt DL, James S, Schneider DJ, Sibbing D, So DYF, Trenk D, Alexopoulos D, et al. International expert consensus on switching platelet P2Y12 receptor-inhibiting therapies. Circulation. 2017. https://doi.org/10.1161/CIRCULATIONAHA.117.031164.Google Scholar
- 37.Cuisset T, Deharo P, Quilici J, Johnson TW, Deffarges S, Bassez C, Bonnet G, Fourcade L, Mouret JP, Lambert M, et al. Benefit of switching dual antiplatelet therapy after acute coronary syndrome: the TOPIC (timing of platelet inhibition after acute coronary syndrome) randomized study. Eur Heart J. 2017;38(41):3070–8.Article PubMed Google Scholar
- 38.De Luca L, D’Ascenzo F, Musumeci G, Saia F, Parodi G, Varbella F, Marchese A, De Servi S, Berti S, Bolognese L. Incidence and outcome of switching of oral platelet P2Y12 receptor inhibitors in patients with acute coronary syndromes undergoing percutaneous coronary intervention: the SCOPE registry. EuroIntervention. 2017;13:459–66.Article PubMed Google Scholar
- 39.Sibbing D, Aradi D, Jacobshagen C, Gross L, Trenk D, Geisler T, Orban M, Hadamitzky M, Merkely B, Kiss RG, et al. Guided de-escalation of antiplatelet treatment in patients with acute coronary syndrome undergoing percutaneous coronary intervention (TROPICAL-ACS): a randomised, open-label, multicentre trial. Lancet. 2017;390(10104):1747–57.CAS Article PubMed Google Scholar
- 40.Angiolillo DJ. Dual antiplatelet therapy guided by platelet function testing. Lancet. 2017;390:1718–20.Article PubMed Google Scholar
Authors’ contributions
Conceived and designed the research: LHC, KM, PS, ARE, JAJ, DJA. Performed the research: FF, FR, LB, AR, MA, DJA. Analyzed the data: FF, FR, DMS, YG. Wrote and edited the paper: LHC, FF, JAJ, DJA. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
DJA reports receiving payments as an individual for: (a) Consulting fee or honorarium from Amgen, Aralez, AstraZeneca, Bayer, Biosensors, Bristol-Myers Squibb, Chiesi, Daiichi-Sankyo, Eli Lilly, Janssen, Merck, PLx Pharma, Pfizer, Sanofi, and The Medicines Company; (b) Participation in review activities from CeloNova and St. Jude Medical. Institutional payments for grants from Amgen, AstraZeneca, Bayer, Biosensors, CeloNova, CSL Behring, Daiichi-Sankyo, Eisai, Eli-Lilly, Gilead, Janssen, Matsutani Chemical Industry Co., Merck, Novartis, Osprey Medical, and Renal Guard Solutions. DJA is recipient of funding from the Scott R. MacKenzie Foundation for human genetic research. LHC receives research funding from Canon Biomedical. None of the other authors have competing interests.
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Study procedures and data collection were approved by the Institutional Review Board at UF Health Jacksonville (IRB201703131), and written informed consent was obtained from all patients included.
Funding
The UF Health Personalized Medicine Program has been funded to date by NIH grants U01 GM074492, U01 HL105198, U01 HG007269, UL1 TR000064 and U54 TR001347 and by substantial institutional support from the University of Florida and its Clinical Translational Science Institute. Spartan Bioscience Inc. (Ottawa, ON) provided the genotyping platforms and kits for testing at the UF Health Jacksonville site. The funding bodies had no role in the design of the study and collection, analysis, or interpretation of the data or in writing the manuscript.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Author notes
- Larisa H. Cavallari and Francesco Franchi contributed equally to this work
Affiliations
- Department of Pharmacotherapy and Translational Research, University of Florida, Gainesville, FL, USALarisa H. Cavallari, D. Max Smith, Yan Gong, Amanda R. Elsey & Julie A. Johnson
- Center for Pharmacogenomics, University of Florida, Gainesville, FL, USALarisa H. Cavallari, D. Max Smith, Yan Gong & Julie A. Johnson
- Clinical and Translational Science Institute, University of Florida, Gainesville, FL, USALarisa H. Cavallari, Amanda R. Elsey & Julie A. Johnson
- Division of Cardiology, Department of Medicine, University of Florida, Jacksonville, FL, USAFrancesco Franchi, Fabiana Rollini, Latonya Been, Andrea Rivas, Malhar Agarwal & Dominick J. Angiolillo
- University of Florida Health Pathology Laboratories, Gainesville, FL, USAKimberly Newsom & Petr Starostik
- Department of Pathology, Immunology and Laboratory Medicine, University of Florida, Gainesville, FL, USAPetr Starostik
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.